- Title
-
The peptide transporter 1a of the zebrafish Danio rerio, an emerging model in nutrigenomics and nutrition research: molecular characterization, functional properties, and expression analysis
- Authors
- Vacca, F., Barca, A., Gomes, A.S., Mazzei, A., Piccinni, B., Cinquetti, R., Del Vecchio, G., Romano, A., Rønnestad, I., Bossi, E., Verri, T.
- Source
- Full text @ Genes Nutr
|
Pairwise alignment between zebrafish PepT1a (Slc15a1a) and PepT1b (Slc15a1b) amino acid sequences obtained by using Clustal Omega and edited using GeneDoc 2.7 software. The predicted conserved PTR2 family proton/oligopeptide symporter signatures (in zebrafish PepT1a, motif 1—PROSITE pattern PS01022—amino acid residues 80–104; and motif 2—PROSITE pattern PS01023—amino acid residues 173–185) are colored in red. In the amino acid sequence, putative transmembrane domains are named I to XII. Weak predicted transmembrane domains (in zebrafish PepT1a, transmembrane domains VIII and X) are colored in gray |
|
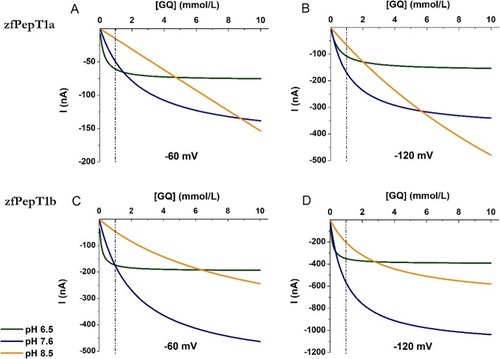
Transport activity and pH dependence of zebrafish PepT1a (Slc15a1a) and PepT1b (Slc15a1b). |
|
Fitting of the Gly-Gln (GQ) transport-associated currents as a function of substrate concentration (from 0.01 to 10 mmol/L) at different pH (pH 6.5 in green, pH 7.6 in blue, and pH 8.5 in orange) for two different membrane potentials: − 60 mV (left) and − 120 mV (right). |
|
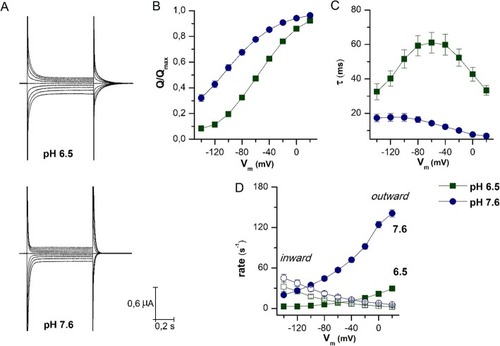
Biophysical parameters of PepT1a. |
|
Expression analysis by RT-PCR on |
|
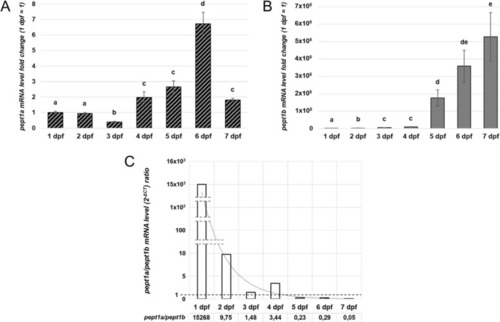
Quantitative expression analysis of zebrafish |