- Title
-
Nrf2 activation attenuates genetic endoplasmic reticulum stress induced by a mutation in the phosphomannomutase 2 gene in zebrafish
- Authors
- Mukaigasa, K., Tsujita, T., Nguyen, V.T., Li, L., Yagi, H., Fuse, Y., Nakajima-Takagi, Y., Kato, K., Yamamoto, M., Kobayashi, M.
- Source
- Full text @ Proc. Natl. Acad. Sci. USA
|
Up-regulated expression of Nrf2 target genes in it768 larvae that have splicing defects in the pmm2 gene. (A) Expression of gstp1 and hmox1a in WT or pmm2it768 larvae injected or not with 1 pmol nrf2aMO. The closed and open arrowheads indicate the gills and liver, respectively. The numbers in each picture indicate the larvae exhibiting strong expression of gstp1 or hmox1a/tested larvae. (B) Nucleotide and deduced amino acid sequences of the major cDNA isolated from WT and pmm2it768 larvae. The red “a” indicates the G-to-A mutation site of the it768 mutant in the zebrafish pmm2 genome. The green “R” indicates the amino acid residue corresponding to Arg141 in human PMM2. EcoRI indicates the EcoRI site only present in the WT cDNA. |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|
|
PERK-dependent activation of Nrf2 in 5-dpf larvae by genetic- and chemical-induced ER stress. The numbers in each picture indicate the larvae exhibiting strong bip expression in the liver (open arrowheads)/tested larvae (A) or strong gstp1 expression in the gills (closed arrowheads)/tested larvae (D–G). (A) bip expression in WT sibling and pmm2it768 larvae. (B) The expression of bip and chop was analyzed by RT-PCR in WT, pmm2it768/+ (m/+), or pmm2it768/it768 (m/m) larvae. The amount of cDNA used for RT-PCR was standardized by the ef1α expression. (C) The relative expression levels of bip and chop to ef1α was evaluated by qPCR in WT, pmm2it768/+ (m/+), or pmm2it768/it768 (m/m) larvae. a and b indicate statistically significant differences (Tukey’s test, P < 0.001; n = 3 for each genotype). (D) Induced expression of gstp1 after 12-h treatment of 5 μg/mL tunicamycin (TM) or 2 μM thapsigargin (TG) in WT AB larvae injected or not with 1 pmol nrf2aMO. (E) Induced expression of gstp1 after 12-h treatment of 1 μM thapsigargin (TG) in WT sibling and nrf2afh318 larvae. (F) gstp1 expression in WT sibling and pmm2it768 larvae injected or not with 1 pmol perkMO. (G) gstp1 expression in WT sibling and perkit312 larvae injected or not with 1 pmol pmm2MO. The graph shows the percentages of gstp1-positive larvae of the indicated genotypes. The numbers of larvae tested are indicated above the bars. Asterisks denote statistical significance (Fisher’s exact test; *P < 0.001). The data were obtained based on the results of the WISH analysis shown in the Upper panels. |
|
The Nrf2-dependent attenuation of the up-regulated ER stress by sulforaphane treatment. The graphs show the percentages of bip-positive 5-dpf larvae of the indicated genotypes and their expression after 12-h treatment of 40 μM sulforaphane (SF). The numbers of larvae tested are indicated above the bars. An asterisk denotes statistical significance (Fisher’s exact test; *P < 0.001). The data were obtained based on the results of the WISH analysis shown in the Right panels. The strength of the bip staining was determined by double-blind scoring. The arrowheads indicate the liver. |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. |
|
The gene responsible for the it768 mutant is pmm2. (A) A physical map of the it768 locus on the zebrafish chromosome 1. The numbers show recombinants identified by a simple sequence-length polymorphism analysis using polymorphic markers. The G-to-A mutation at position 1 of the 5′ splice site in the intron 5 was found in the mutant allele. (B) A phylogenetic tree constructed by the neighbor-joining method for the full-length amino acid sequences of Pmm proteins. (C) gstp1 expression at 5 dpf in WT or pmm2it768 larvae injected or not with 1 pmol each of the indicated morpholino oligonucleotides. The closed and open arrowheads indicate the gills and liver, respectively. (D) gstp1 expression at 5 dpf in WT or pmm2it768 larvae injected or not with mRNA encoding zebrafish Pmm2 protein. (E) Results of the RT-PCR analysis using total RNA isolated from WT, pmm2it768/+ (m/+), or pmm2it768/it768 (m/m) larvae at 5 dpf and primers designed to amplify partial cDNA corresponding to the exon 5–intron 5–exon 6 region. EcoRI cut the cDNA from the normally spliced mRNA. (F) Results of the cDNA analysis prepared from total RNA of whole bodies in WT or pmm2it768 larvae at the indicated dpf. The numbers of the sequenced cDNA and the percentages of the indicated splicing types are shown. The asterisks indicate stop codons. (G) Amino acid sequences corresponding to exons 5 and 6 of the vertebrate and invertebrate Pmm proteins. The highlighted amino acid residues are highly conserved among eukaryotes. at, Arabidopsis thaliana; c, chicken; ce, Caenorhabditis elegans; d, Drosophila melanogaster; h, human; m, mouse; sc, Saccharomyces cerevisiae; x, Xenopus laevis; z, zebrafish. |
|
pmm2 expression during the embryonic and larval stages. (A) Ubiquitous expression of pmm2 at the indicated developmental stages in WT AB embryos. (B) pmm2 expression at the indicated developmental stages in WT or pmm2it768 homozygous mutants. The expression in the gills (closed arrowheads) and liver (open arrowheads) was up-regulated in pmm2it768 homozygous larvae. PHENOTYPE:
|
|
The expression of gstp1 and bip in WT AB larvae treated with ER stress inducers. (A) Induced expression of gstp1 in 5-dpf larvae treated with 5 μg/mL tunicamycin (TM), 2 μM thapsigargin (TG), or 100 μM diethyl maleate (DEM) in the indicated treatment time. The closed and open arrowheads indicate the gills and liver, respectively. (B) Induced expression of bip in 5-dpf larvae injected or not with 1 pmol nrf2aMO after 12-h treatment of 5 μg/mL tunicamycin (TM) or 100 μM diethyl maleate (DEM). The numbers indicate the larvae exhibiting strong bip expression in the liver (open arrowheads)/tested larvae. |
|
Loss-of-function analyses of zebrafish PERK. (A) A phylogenetic tree constructed by the neighbor-joining method for the full-length amino acid sequences of PERK proteins. c, chicken; ce, C. elegans; d, D. melanogaster; h, human; m, mouse; xt, Xenopus tropicalis; z, zebrafish. (B) The expression of perk, chop, and bip analyzed by RT-PCR using total RNA from 8-h–postfertilization embryos injected or not with 1 pmol perkMO. s, spliced form; us, unspliced form. The amount of cDNA used for RT-PCR was standardized by the ef1α expression. (C) Induced expression of gstp1 after 12-h treatment of 5 μg/mL tunicamycin (TM), 2 μM thapsigargin (TG), or 100 μM diethyl maleate (DEM) in WT AB larvae injected or not with 1 pmol perkMO. The numbers in each picture indicate the positive larvae/tested larvae. (D) The deletion in exon 13 of perk in the it312-mutant genome and schematic representations of the WT and mutant proteins. A dotted bar indicates an artificial 66-aa C-terminal region generated by the frame shift mutation. IRE1-like, IRE1-like domain; Kinase, kinase domain; SP, signal peptide region; TM, transmembrane domain. |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. |
|
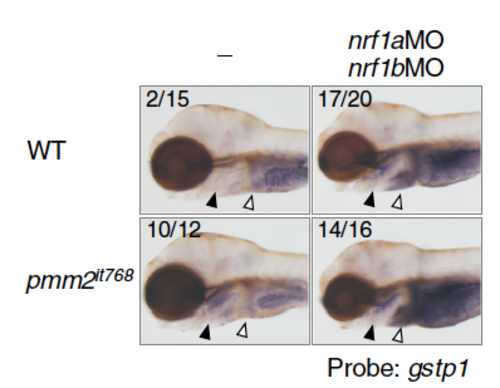
The expression of gstp1 in the gills (closed arrowheads) and liver (open arrowheads) analyzed in nrf1a- and nrf1b-double morphants. gstp1 expression in WT sibling and pmm2it768 larvae injected or not with 0.5 pmol each of nrf1aMO and nrf1bMO. The numbers indicate the larvae exhibiting strong gstp1 expression in the gills/tested larvae. |