- Title
-
Inhibition of signaling between human CXCR4 and zebrafish ligands by the small molecule IT1t impairs the formation of triple-negative breast cancer early metastases in a zebrafish xenograft model
- Authors
- Tulotta, C., Stefanescu, C., Beletkaia, E., Bussmann, J., Tarbashevich, K., Schmidt, T., Snaar-Jagalska, B.E.
- Source
- Full text @ Dis. Model. Mech.
|
CXCR4 expression levels correlate with metastatic potential in a zebrafish xenotransplantation model. The bone clone (MDA-MB-231-B) expressed higher levels of CXCR4 mRNA (A) and lower levels of CXCR7 mRNA (B), compared to the parental cell line MDA-MB-231, originated from metastatic triple-negative breast cancer (TNBC) [unpaired t-test, (A) **P=0.0016, (B) **P=0.0019]. Upon engraftment into the duct of Cuvier (C,C′ red arrow) of 2-dpf zebrafish embryos (C′), MDA-MB-231 circulated in the vascular system (D), in a comparable manner to MDA-MB-231-B (E). Arrowhead in C represents the site of injection. CHT, caudal hematopoietic tissue. (F-J) Tumor cells disseminated throughout the embryo, in the head (F-H), the eye (H), the trunk and the tail (I,J), and extended filopodia at vessel branching points (I, arrowhead). BA, basilar artery; BAs, branchial arches; CV, caudal vein; DA, dorsal aorta; DLAV, dorsal longitudinal anastomotic vessel; ISV, intersegmental vessel. (K-P′) Over time, a weaker phenotype was detectable for the MDA-MB-231 cell line (K-M,K′-M′), whereas evident secondary tumor mass formation, extravasation and tail fin invasion persisted when MDA-MB-231-B cells were implanted (N-P,N′-P′). Arrows in O′ and P′ indicate invasive cancer cells that are not in contact with the endothelium and are found in the tail fin tissue, after extravasation. Images were acquired using a Leica TCS SPE confocal microscope with an HC PL FLUOTAR 10× DRY objective (0.30 N.A.) in panel C and with an HC APO 20× DRY objective (0.7 N.A.) in panel H. All other images were acquired using a Leica MZ16FA fluorescent microscope coupled to a DFC420C camera. Scale bars: 50µm. Phenotype assessment was carried out at 1, 2 and 4dpi, evaluating the ability of both cell lines to form a secondary tumor mass, to extravasate and to invade the surrounding tail fin. Images are representative of embryos injected with MDA-MB-231 and number of individuals was n=51 (5hpi) (D), 45 (1dpi) (K), 44 (2dpi) (L) and 25 (4dpi) (M) or with MDA-MB-231-B and number of individuals was n=44 (5hpi) (E), 42 (1dpi) (N), 36 (2dpi) (O) and 34 (4dpi) (P). Percentages relative to tumor mass (TM), tumor extravasation (TE) and tumor invasion (TI) are reported for each stage, for both MDA-MB-231 and MDA-MB-231-B cell lines. |
|
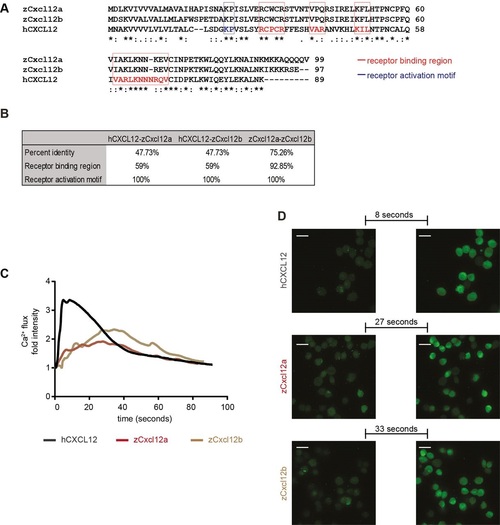
Zebrafish Cxcl12 ligands activate CXCR4 signaling in human cancer cells. Human CXCL12 was aligned to the zebrafish homologs using ClustalW (A). Amino acid residues were conserved in the receptor binding region and activation motif of CXCL12/Cxcl12 chemokines (A,B). Asterisks (*) represent fully conserved residues; colons (:) and periods (.) indicate positions at which residues share strong or weak similarity, respectively. (C,D) Human CXCL12-α and zebrafish ligands Cxcl12a and Cxcl12b induced calcium flux in MDA-MB-231-B cells, as detected by increased fluorescence intensity. The human ligand initiated an immediate response that extinguished rapidly (31s to register half fluorescence intensity after the highest response), whereas the zebrafish ligands triggered a slower and prolonged signal induction (>55s for zCxcl12a and >52s for zCxcl12b to register half fluorescence intensity). (C) The fold intensity increase is calculated by normalization on fluorescence intensity correspondent to signal before response activation. (D) Frames show intensity before signaling activation was triggered by each ligand and at the highest peak of response. The time length to reach the strongest response is indicated. |
|
Human CXCL12 triggers zebrafish macrophage migration in a Cxcr4-dependent manner. (A) Scheme of a 30- to 32-hpf embryo and injection site are shown. (B,C) Zebrafish macrophages were found to be responsive to human CXCL12 (0.4mg/ml) 3h after injection into the hindbrain ventricle (HBV) of 30- to 32-hpf embryos. No increase in macrophage number occurred when a mock solution (water) was inoculated. Zebrafish Cxcl11aa (1.5mg/ml) was used as a positive control (C). Data in C are pooled observations from two independent experiments (n=55 in mock; n=48 in hCXCL12; n=57 in zCxcl11aa). (D,E) Macrophages were recruited by human CXCL12 in a Cxcr4-dependent manner: a higher number (43%) of L-P+/TSA- cells was found in the HBV compared to the mock-injected group in wild-type (wt) siblings (D,E), whereas no differences were detected in the cxcr4b-/- (ody) mutants (E). In B, mCherry-expressing macrophages are recruited by hCXCL12 as in D, where L-P staining combined to TSA detection is used to distinguish macrophages (L-P+/TSA-) from neutrophils (L-P+/TSA+). ****P<0.0001, ns P>0.05 one-way ANOVA, Bonferroni post-hoc test. Data in E are pooled observations from five independent experiments (n=171 in mock/wt; n=180 in hCXCL12/wt; n=139 in mock/ody; n=160 in hCXCL12/ody). |
|
CXCR4-expressing TNBC cells fail to initiate metastatic events in cxcl12a- and cxcl12b-null zebrafish mutants. (A) No differences in tumor cell invasion were found in the medusa (cxcl12a-/-) mutants, compared to wild-type (wt) siblings. (B,C) Breast cancer cells failed to form micrometastases in 4-dpi zebrafish embryos deficient for both cxcl12a and cxcl12b ligands, whereas tumor invasion (B) and tumor burden (C) occurred in the cxcl12a-/-/cxcl12b+/+ and cxcl12a-/-/cxcl12b+/- siblings. Unpaired t-test: (A) ns, P>0.05 (wt: n=73; medusa: n=66), (B) **P=0.0012 and (C) **P=0.0033 (n=11 in each group). Graphs in A-C are cumulative of two independent experiments. (D,E) Top panels: MDA-MB-231-B breast cancer cells were highly invasive in the siblings, whereas few tumor cells remained in the metastatic region in zebrafish that were mutant for both ligands. Images were acquired using a Leica MZ16FA fluorescent microscope coupled to a DFC420C camera. Bottom panels: vessel connections are compared to distinguish siblings and double mutants: the hypobranchial arteries (HA), indicated by asterisks, failed to connect to the mandibular arch (AA1) in the cxcl12a-/-/cxcl12b-/- larvae. Images were acquired using a Leica TCS SPE confocal microscope with an HC PL FLUOTAR 10× DRY objective (0.30 N.A.). Scale bars: 50µm. (F) CXCL12 expression is lower in the MDA-MB-231-B compared to MCF-7 breast cancer cell line. Unpaired t-test, with Welch′s correction. **P=0.006. qPCR was performed on two biological replicates. |
|
The CXCR4 antagonist IT1t reduces metastatic tumor burden in vivo. IT1t (20µM) was applied into the cell medium for 24h prior to engraftment in zebrafish embryos to antagonize CXCR4 receptor activation (A). The percentage of live cells after treatment was not significantly different than in the control condition (three independent experiments) (B). The metabolic activity, readout of cell growth, was not significantly affected when increasing concentrations of IT1t were used (C). Pre-treatment in vitro caused a reduction in tumor burden at the secondary site in vivo, both at 2dpi (D) and 4dpi (E). Cancer cell burden increased over time from 2 to 4dpi in the control group, whereas it remained at comparable levels upon treatment (F). Data set in F is obtained by using the same data points as shown in D and E. Number of larvae is n=64 (DMSO) and n=75 (IT1t) at 2dpi and n=59 (DMSO) and n=56 (IT1t) at 4dpi. (G) Effect of CXCR4 inhibition on tumor burden over time is shown. Scale bars: 50µm. Data are mean±s.e.m. from two independent experiments. Statistical analysis: two-tailed, unpaired t-test and ANOVA with Bonferroni post-hoc test for datasets with two or more groups respectively. ****P<0.0001; **P=0.005; ns, P>0.05. |
|
Ca2+ efflux in the parental line MDA-MB-231. Ca2+ mobilization was assessed upon stimulation with 100 and 300 nM hCXCL12. (A) After addition (32nd second, black arrow) of 100 nM hCXCL12, no response was detected in MDA-MB-231. The same sample was subjected to stimulation with higher concentration (300 nM). In this case, the chemokine addition (8th second, grey arrow) resulted in an initiation of response (20th second, blue arrow), with the highest signal intensity registered 1 minute later (81st second, green arrow). The red arrow indicates the expected time of highest signal intensity (8 seconds after signal initiation), which was observed upon stimulation of MDA-MB-231-B with 100 nM hCXCL12 (Figure 2D). (B) Signal intensity at ligand addition and 60 seconds after stimulation. No difference in signal intensity was observed with 100 nM hCXCL12, while fluorescence increase was registered with 300 nM hCXCL12. Scale bar: 50 µm. |
|
Cxcr4b does not affect macrophage basal motility and total number. (A) Macrophage tracks in a wild-type Tg(mpeg1:EGFP) gl22 sibling. (B) Macrophage tracks in a cxcr4b -/- Tg(mpeg1:EGFP) gl22 sibling. (A′, B′) Bright field images of (A) and (B), respectively show the orientation of the tail fins. Scale bar: 50 µm. (C) Average speed and (D) cumulative distance of macrophages do not differ in WT and Ody larvae at 4 dpf. Statistical significance is assessed using unpaired t-test, ns p> 0.05. (E) Total number of macrophages detected by immunohistochemistry is the same in Ody and WT siblings at 2 dpf. Statistical significance is assessed using Mann-Whitney test, ns p> 0.05. WT: n=57, Ody: n=55. |
|
Lack of functional ligand affects MDA-MB-231-B cell positioning. (A) MDA-MB-231-B cell localization in the head region is reduced in cxcl12a -/- mutants, in particular in branchial arches (B, C) and basal artery (C). Localization in the intersegmental vessels is also diminished (D). |
|
Micrometastasis formation occurs in cxcl12 single ligand mutants. Engraftment of MDA-MB-231-B cell line resulted in micrometastasis formation in the tail fin region, characterized by invading single cells in both the cxcl12b -/- mutant and wt siblings at 2 and 4 dpi (A) and tumor burden in wt, cxcl12a -/- and cxcl12b -/- larvae at 2 dpi (B and C). The trend was not statistically significant (One-way ANOVA and Bonferroni post hoc test). Number of larvae in (A): n=30 (wt 2 dpi), n=20 (cxcl12b -/- 2 dpi), n=19 (wt 4 dpi) and n=15 (cxcl12b -/- 4 dpi). Number of larvae in (B): n= 23 (wt), n=27 (cxcl12a -/-) and n=27 (cxcl12b -/-). Scale bars: 50 µm. |

Unillustrated author statements PHENOTYPE:
|