- Title
-
Matrix metalloproteinase 9 modulates collagen matrices and wound repair
- Authors
- LeBert, D.C., Squirrell, J.M., Rindy, J., Broadbridge, E., Lui, Y., Zakrzewska, A., Eliceiri, K.W., Meijer, A.H., Huttenlocher, A.
- Source
- Full text @ Development
|
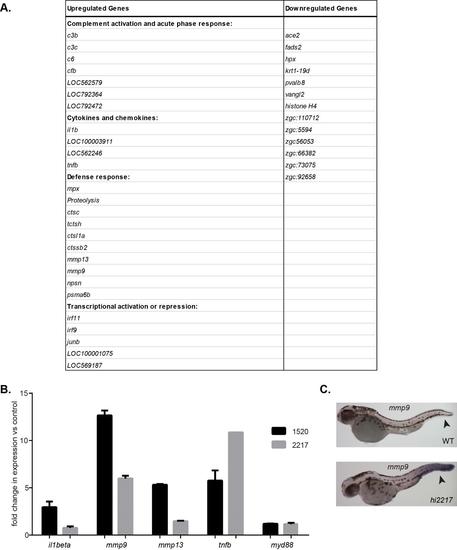
Gene expression profiling reveals elevated inflammatory gene expression, including mmp9, in chronic inflammation mutants. (A) Microarray analysis of inflammation mutants, hi1520 and hi2217, revealed overlapping upregulation of 150 genes and downregulation of 16 genes compared with WT siblings. (B) Upregulation of mmp9 was confirmed by qRT-PCR in the hi2217 and hi1520 mutants and the hai1 morphants. (C) The mutants are characterized by epithelial extrusions and abnormal epithelium development (arrows). Data pooled from experiments performed in triplicate. *P<0.05. Scale bar: 200 µm. PHENOTYPE:
|
|
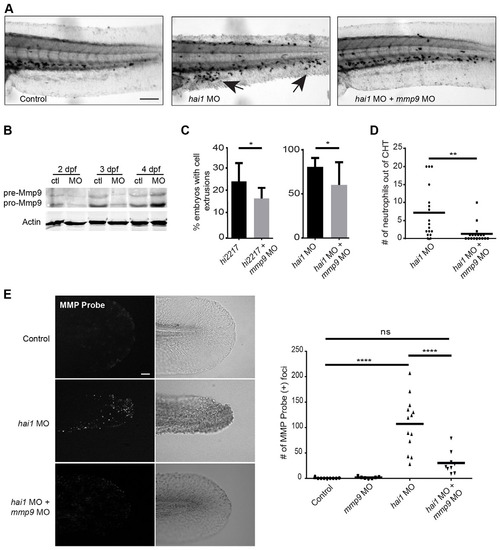
Inflammatory phenotypes associated with hai1 mutants are partially rescued by knockdown of mmp9. (A) Sudan Black staining of the hai1 mutants show increased neutrophilic infiltration of the epithelium (arrows), a phenotype that could be rescued by mmp9 knockdown. (B) Western blot analysis of mmp9 MO1. (C) Morpholino knockdown of mmp9 (MO1) decreased the proportion of hi2217 and hai1 morphants displaying epithelial extrusions and (D) decreased the number of neutrophils infiltrating the epithelium in the hai1 morphants. (E) MMPSense showed hyper-activation of MMPs in the hai1 morphants. Hyper-activation of MMPs in the hai1 morphants could be partially rescued by the knockdown of mmp9 expression (MO1). *P<0.05, **P<0.01 and ****P<0.0001. Scale bars: 300µm in A; 40µm in E. C and D represent data from a single experiment performed in triplicate. PHENOTYPE:
|
|
SHG imaging shows altered collagen alignment in hai1 morphants that is partially rescued by mmp9 depletion. (A) SHG imaging of type I/III collagen. Caudal fins were excited with an 890′w laser and collagen was detected at 445′w. (B) Representative z-projected images of stitched multiple ROI SHG z-stacks. The hai1 morphants display irregular collagen alignment. (C) In order to quantify alignment, SHG images illustrate scoring scheme, ranking severity of collagen mis-alignment for analysis from 1 to 3. (D) The hai1 morphants display a significant decrease in collagen alignment that could be partially rescued upon knockdown of mmp9 expression (MO1). (E) IHC of type IV collagen in the transgenic zebrafish Tg(krt4:tdTom). Epithelial extrusions were observed (arrows) in the hai1 morphants. Knockdown of mmp9 resulted in fewer observed extrusions. *P<0.05 and ***P<0.001. Scale bars: 50µm in B,C; 20µm in E. D represents the data from experiments performed in quadruplicate and scored by an individual, single-blind analyzer. D represents pooling with experimental numbers for Control MO=18, mmp9 MO=18, hai1 MO=20, hai1 MO+mmp9 MO=20. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Acute wounding induces NFκB-dependent expression of mmp9. (A) An increased expression of mmp9 is observed by 6h post wounding (hpw) in the 2-dpf embryo. (B) A visible increase in NFκB activation is seen by 6hpw using the transgenic Tg(NFκB:gfp). The star denotes neuromast cells expressing high levels of gfp independent of wounding. The arrow indicates the edge of the wounded caudal fin and the site of increased NFκB activation. (C) Ratiometric image analysis performed in a cross of Tg(NFκB:gfp) and Tg(krt4:tdTom). (D) Early inhibition of the NFκB pathway with 30µM withaferin A resulted in a significant reduction in regeneration at 3dpw but had no effect on developmental fin length. (E) Early inhibition of the NFκB pathway resulted in an abrogation of the increased mmp9 expression in the Tg(krt4:l10a-gfp) TRAP line. *P<0.05, **P<0.01 and ****P<0.0001. Scale bars: 40µm in B,C; 100 µm in D. A represents data combined from experiments performed in triplicate; D represents data combined from experiments performed in quadruplicate and normalized to unwounded treatments. |
|
SHG imaging reveals a defect in collagen thickening in the caudal fin of mmp9 morphants at 2days post amputation. (A) Amputation of the larval caudal fin results in a thickening of type I/III collagen fibers by 2dpw (top). Fiber thickening does not appear to occur in the mmp9 morphants (MO1) by 2dpw (bottom). (B) Scoring system for fiber width, from 1 to 3 (top). A significant defect in collagen fiber thickness is seen in the mmp9 morphants at 2dpw (bottom). (C) Schematic illustrating the quantification of SHG images. All fibers were measured and the number of fibers above a given threshold width (i.e. 7 pixels) can be determined, see Materials and Methods. (D) Quantification of the number of fibers >7 pixels in diameter per caudal fin validates a defect in collagen fiber thickening in the mmp9 morphants at 2dpw. **P<0.01, ***P<0.001 and ****P<0.0001. Scoring in B from a single-blind analyzer from pooled experiments performed in triplicate. Data in D are from experiments performed in quadruplicate and scored by a single-blind analyzer. Scale bars: 50µm in A; 20µm in B. Experimental numbers for B and D: no wound Control MO=21, no wound mmp9 MO=25, wounded Control MO=31, wounded mmp9 MO=32. PHENOTYPE:
|
|
Mmp9 regulates caudal fin wound healing following amputation. (A) Morpholino knockdown of mmp9 (MO1) causes a regenerative defect at 3dpw without influencing developmental fin length. (B) Co-injection of mmp9 MO2 and zebrafish mmp9 RNA (125ng/µl) rescues the healing defect at 3dpw (5dpf). (C) Schematic of two-site CRISPR-Cas9 targeting of mmp9 (top). Half-stars indicate CRISPR sites and arrows represent primer binding for screening. PCR amplification results in the presence of a ~400bp amplicon indicated by the star, with the ctl lane representing a housekeeping gene to ensure the presence of gDNA (bottom). (D) Mosaic F0 larvae did not have altered fin lengths at 5dpf but displayed a significant defect in caudal fin wound healing. *P<0.05, **P<0.01, ***P<0.001 and ****P<0.0001. Graphs represent a single experiment performed in triplicate. Scale bars: 100µm in A,D. PHENOTYPE:
|
|
Inflammation-related gene expression changes are observed in the chronic inflammation mutants hi2217 and hi1520. A) Microarray analysis revealed many inflammationrelated genes that were differentially expressed in both inflammation mutants. For a complete list, see GSE28110. B) Confirmation of selected microarray target genes by qRT-PCR confirmed mmp9 to be highly overexpressed in both of the inflammation mutants. C) In situ hybridization of mmp9 (arrowhead) in the hi2217 mutants. For qRT-PCR expression analysis, expression was normalized to ef1α expression. |
|
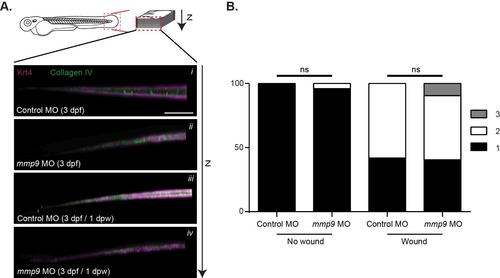
No defects in collagen type IV (IHC) or alignment of collagen fibers are observed at 2 dpw. A) Antibody labeling (IHC) of type IV collagen did not yield obvious differences between the different conditions. B) Blind analysis for collagen alignment (SHG) at 2 dpw indicated a nonsignificant increase in collagen mis-alignment in both control and mmp9 morphant caudal-fins. Graph represents data from experiments performed in quadruplicate and scored by an individual, single-blind analyzer. Scale bar in A represents 50 µm. Graph represents pooling with experimental numbers for no wound Control MO = 21, no wound mmp9 MO = 25, wounded Control MO = 31, wounded mmp9 MO = 32. |