- Title
-
Functionally conserved cis-regulatory elements of COL18A1 identified through zebrafish transgenesis
- Authors
- Kague, E., Bessling, S.L., Lee, J., Hu, G., Passos-Bueno, M.R., and Fisher, S.
- Source
- Full text @ Dev. Biol.
|
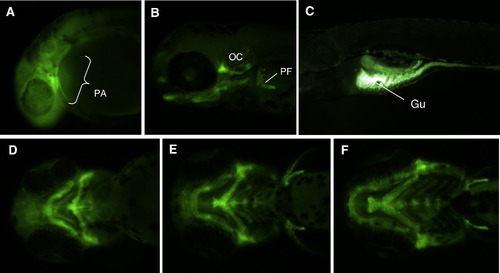
Zebrafish col18a1 displays a complex and dynamic development expression pattern. Expression of col18a1 was detected by whole embryo in situ hybridization at 24 hpf (A); 2 dpf (B); and 3 dpf (C–E). Dynamic expression was seen in neutral structures including spinal cord (A) and tectum (B). Expression was also seen in pronephric duet and pronephros (A, E); myospetum (A); epidermis (A, B); otic vesicle (B); pharyngeal arches (B); craniofacial cartilages and blood vessels (C); retinal pigment epithelium (D); pectoral fins (E); and gut endothelium (E). Abbreviations used in all figures: BV (blood vessels), Ca (cartilage), FB (forebrain), Gu (gut endothelium), My (myoseptum), OV (otic vesicle), PA (pharyngeal arches), PD (pronephric duct), PF (pectoral fin), Pr (pronephros), Re (retina), RPE (retinal pigment epithelium), SC (spinal cord), Te (tectum). EXPRESSION / LABELING:
|
|
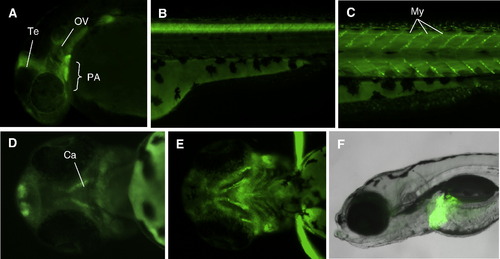
Overlapping upstream sequences regulate similar patterns of EGFP expression in zebrafish embryos. Embryos transgenic for either CNSv -97.8 or CNSp -96.9 constructs had EGFP expression at 24 hpf in forebrain, retina, tectum, otic vesicle, and pronephric duct (A–D), and transiently in somites (A, C). At 3 dpf, EGFP was expressed in cartilage of the head, pectoral fins, and otic vesicle (E, F). EXPRESSION / LABELING:
|
|
An upstream enhancer sequence regulates expression in brain, cartilage, and gut. Embryos transgenic for CNSv -78.7 constructs had EGFP expression at 24 hpf in the brain and pharyngeal arches (A). Later expression was seen in craniofacial, otic capsule, and pectoral fin cartilages at 3 dpf (B, D), 4 dpf (E) and 5 dpf (F). EGFP expression was also seen in gut endothelium at 5 dpf (C). |
|
Blood vessels expression of COL18A1 is mediated by an intronic enhancer element. Embryos transgenic for the CNSp +47.8 constructs displayed EGFP expression at 1 (A) and 2 (B) dpf in the pharyngeal arches and heart (A, B), and at 3 dpf in blood vessels and cartilage of the pharyngeal arches (C, D), in the retina pigment epithelium (D), and in the dorsal aorta and other blood vessels of the trunk (E, F). EXPRESSION / LABELING:
|
|
A second intronic enhancer mediates expression in liver and other tissues. CNSv +64.8 regulated EGFP expression at 24 hpf in tectum, otic vessel, pharyngeal arches (A), and spinal cord (B), and at 2 dpf in myoseptum (C). Expression was also seen in craniofacial and pectoral fin cartilage at 2 (D) and 3 (E) dpf, and in liver at 5 dpf (F). |
|
Identification of an orthologous zebrafish enhancer sequence showing functional overlap with CNSp +47.8 A) The intergenic sequence of COL18A1 encompassing CNSp +47.8 was compared to orthologous regions from mouse, stickleback, and zebrafish, using Shuffle-Lagan. The turquoise line at top indicates the extent of the human enhancer element, and blue the nearby coding exon, yellow blocks on the second line show the location of repetitive sequences. On the tracks for each species, colored peaks are those meeting the criteria for conservation: turquoise aligning with the enhancer, red with other non-coding sequence, and blue with the exon. The analysis revealed alignment between the human enhancer and zebrafish non-coding sequence; the fragment indicated by the red bar was cloned as described in the text. Note that the larger alignment peak adjacent to the cloned sequenced falls within repetitive sequence in both the human and zebrafish genomes. The zebrafish construct, zCNS +88, regulated EGFP expression in multiple embryonic tissues, including B) heart and otic vesicle; C) aorta and floorplate (asterisk); cartilage (D, E); blood vessels of the pharyngeal arches (E, F); and cranial blood vessels (arrowheads in G). |
|
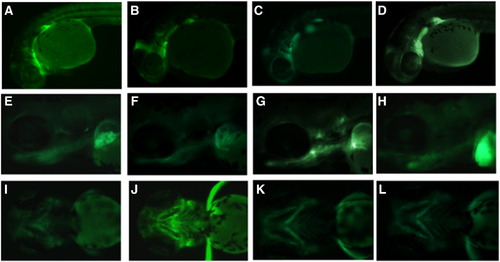
Four independent transgenic lines of CNS -97.8 show substantially similar expression. EGFP expression is shown in the heads (A–D) and in the trunks (E–H) of 1 dpf embryos from four different founders. |
|
Four independent transgenic of CNS +64.8 shows substantially similar expression. (A–D) Lateral views of 1 dpf embryos from four different founders show prominent expression in tectum, otic vesicle, and pharyngeal arches. At 3 dpf (E–H) and at 4 dpf (I–L), continued expression in cartilages of pharyngeal arches and pectoral fine is observed an all four lines. |
|
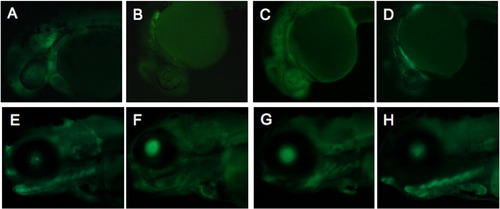
Three independent lines of CNS +47.8 demonstrate some variability in expression pattern. Embryos from three founders are shown at 1 dpf embryos (A–C), 2 dpf (D–F), and 4 dpf (G–I). Some variability of EGFP expression among the lines is apparent; for example, the first line has stronger and more persistent cartilage expression (G), but most embryos display and the dominant pattern of expression in cartilage, heart, and blood vessels. |
|
Four independent transgenic lines for zCNS +88 show substantial similarity. Embryos from four founders display essentially similar patterns of expression at 1 dpf (A–D) and 4 dpf (E–F), including expression in heart and vasculature. |
Reprinted from Developmental Biology, 337(2), Kague, E., Bessling, S.L., Lee, J., Hu, G., Passos-Bueno, M.R., and Fisher, S., Functionally conserved cis-regulatory elements of COL18A1 identified through zebrafish transgenesis, 496-505, Copyright (2010) with permission from Elsevier. Full text @ Dev. Biol.