- Title
-
Control of Dead end localization and activity – Implications for the function of the protein in antagonizing miRNA function
- Authors
- Slanchev, K., Stebler, J., Goudarzi, M., Cojocaru, V., Weidinger, G., and Raz, E.
- Source
- Full text @ Mech. Dev.
|
Dead end 3′UTR is responsible for targeting of dnd mRNA to the PGCs. (A, B) 1-cell stage embryos injected with GFP mRNA fused to Dnd 3′UTR were subjected to in situ hybridization with GFP anti-sense probe. GFP mRNA is restricted to the PGCs only by the dnd 3′UTR (A), in contrast to the globin driven ubiquitous mRNA expression (B). Accordingly, the GFP protein produced by the fusion mRNA follows the same pattern and is detected by fluorescent microscopy at 24hpf only in the PGCs in the case of dnd UTR (C) or ubiquitously as for globin UTR (D). |
|
Deletions of parts of Dead end protein reveal domains important for protein localization and function. (A) Schematic drawing of Dead end deletion constructs. (B) Intracellular localization of the various Dnd-deletion fusion proteins detected by immunohistochemistry with anti-HA antibody (Green) in comparison with the position of the GCGs reported by anti-vasa antibody staining (Red). (C) A chart depicting the ability of the various deletions constructs to supplement for wild type Dead end. The values represent the number of rescued PGCs in embryos co-injected with a specific Dnd truncated protein and dnd MO, normalized to the PGCs rescued by a wild type dnd mRNA injection (see the text for details). PHENOTYPE:
|
|
Dead end shuffles between the nucleus and the cytoplasm. (A) Dnd subcellular localization was changed from cytoplasmic to nuclear when amino acids Y72, F89 and N94 were altered. (B) Dead end protein was trapped in the nucleus when the Exportin 1 mediated nucleocytoplasmic export was blocked by leptomycin B (LMB). The localization of Dead end (green) was compared to that of Vasa protein (red), which labels the perinuclear granules in the control embryos treated with DMSO only (upper panel) and LMB treated embryos (lower panel). |
|
Sub-cellular localization of Dnd versions in PGCs and somatic cells. (A) Similar to other loss of function mutations within the RRM (see Fig. 5), the Y104C mutated Dnd is localized to the nucleus. In contrast, Dnd with loss of function mutation K200T, which resides outside the RNA binding domain, was localized to the germ-cell granules. (B) Over-expression of Dead end in somatic cells using globin 3′UTR. Within the PGCs, outlined in red using membrane bound mCherry under the control of nanos1 3′UTR, Dnd is localized to the germ-cell granules. In the somatic cells, Dnd is ubiquitously distributed within the cell. |
|
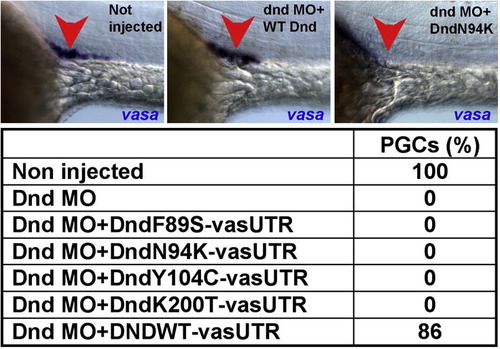
Mutated Dnd proteins translated of RNAs containing the Vasa 3′UTR fail to support PGC survival in dnd MO injected embryos. Embryos were injected simultaneously with dnd MO and mRNA of Dnd mutants, fixed at 24hpf and subjected to in situ hybridization with vasa anti sense probe. Similar to the nanos1 3′UTR results, RNAs encoding Dnd mutated in certain critical amino acids failed to reverse the effect of the antisense oligonucleotides. |
Reprinted from Mechanisms of Development, 126(3-4), Slanchev, K., Stebler, J., Goudarzi, M., Cojocaru, V., Weidinger, G., and Raz, E., Control of Dead end localization and activity – Implications for the function of the protein in antagonizing miRNA function, 270-277, Copyright (2009) with permission from Elsevier. Full text @ Mech. Dev.