- Title
-
Many ribosomal protein genes are cancer genes in zebrafish
- Authors
- Amsterdam, A., Sadler, K.C., Lai, K., Farrington, S., Bronson, R.T., Lees, J.A., and Hopkins, N.
- Source
- Full text @ PLoS Biol.
|
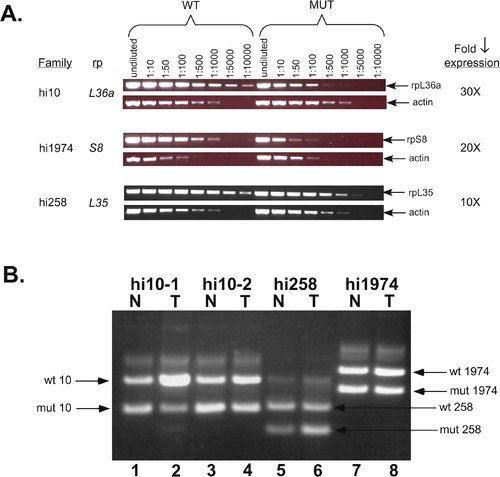
Figure 4. RP Genes Appear to Be Haploinsufficient Tumor Suppressors (A) RP mutations decrease the amount of RP gene expression. RNA was prepared from 3-d-old homozygous mutant embryos and their wild-type siblings, and serial dilutions of first strand cDNA were used as templates for PCR. The decrease in expression in the mutants can be determined by the difference in the dilution between wild type and mutant where the PCR product amount diminishes. The actin control shows that the total amount of mRNA was the same between samples. (B) LOH is not observed in RP mutant tumors. DNA was prepared from tumors (T) and normal tissue (N) from the same fish, and PCR was conducted with three primers that show the presence or absence of both the insert-bearing (mutant) and wild-type chromosomes. In each case, the upper band is the wild-type chromosome and the lower band is the insert-bearing one. hi10 fish #1 normal (lane 1), tumor (lane 2); hi10 fish #2 normal (lane 3), tumor (lane 4); hi258 fish normal (lane 5), tumor (lane 6); hi1974 fish normal (lane 7), tumor (lane 8). EXPRESSION / LABELING:
|
|
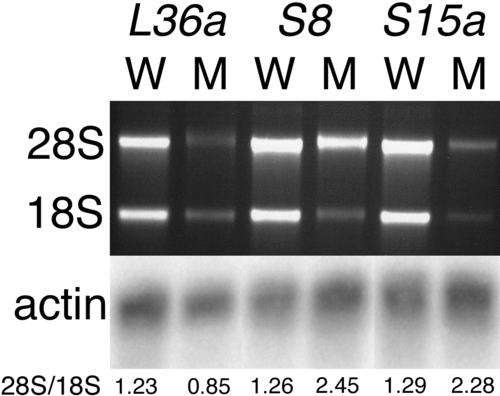
Figure 5. Ribosomal RNA Levels Are Reduced in RP Mutants RNA was prepared from 3-d-old homozygous mutant embryos or their wild-type siblings from lines hi10 (L36a), hi1974 (S8), and hi2649 (S15a), and RNA content was visualized by electrophoresis and ethidium bromide staining. The ratio of 28S/18S as determined by densitometry is shown below each lane. Note that L36a mutants show a preferential loss of the 28S band by 1.5-fold, while S8 and S15a mutants show a preferential loss of the 18S band by 1.9- and 1.8-fold, respectively. These RNAs were also northern blotted and probed for beta actin as an mRNA content control. |