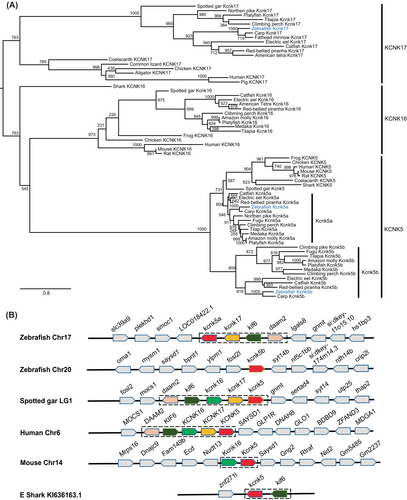
Fig. 8 The kcnk5 gene was duplicated in teleosts, including zebrafish. (A) Molecular phylogenetic tree generated using maximum likelihood analysis on vertebrate tandem of pore domains in a weak inward rectifying K+-related alkaline pH-activated K+ (TALK) channel proteins as obtained with Jones–Taylor–Thornton model plus gamma distribution. Numbers at each node denote bootstrap values based on 1000 replicates. Branch lengths are proportional to expected replacements per site. The zebrafish channels were highlighted in blue. The TALK channels, KCNK5, KCNK16, and KCNK17 formed distinct clades. The Kcnk5a and Kcnk5b formed two subgroups within the KCNK5 clade. (B) The synteny of KCNK5 in five representative vertebrate species. The illustration of the genes and their sizes are not proportional to the length of the distances between genes. KCNK5 was highlighted in red. The conserved synteny (daam2–kif6–kcnk16–kcnk17–kcnk5) was boxed with dashed lines. This synteny in elephant shark is short due to limited available genomic information.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Dev. Dyn.