Fig. 5
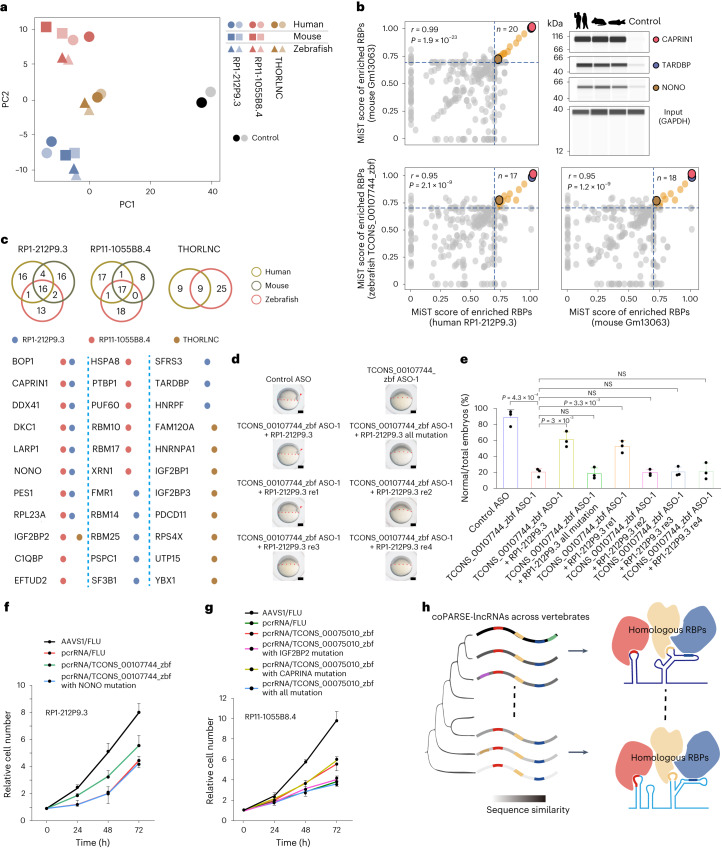
Fig. 5: Identification and functional analysis of the RBP interactome for two coPARSE-lncRNAs., PCA of MS data for HeLa cell lysates pulled down for the indicated human coPARSE-lncRNAs and the predicted mouse and zebrafish homologs. The control samples are based on luciferase transcript segments.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Nat. Genet.