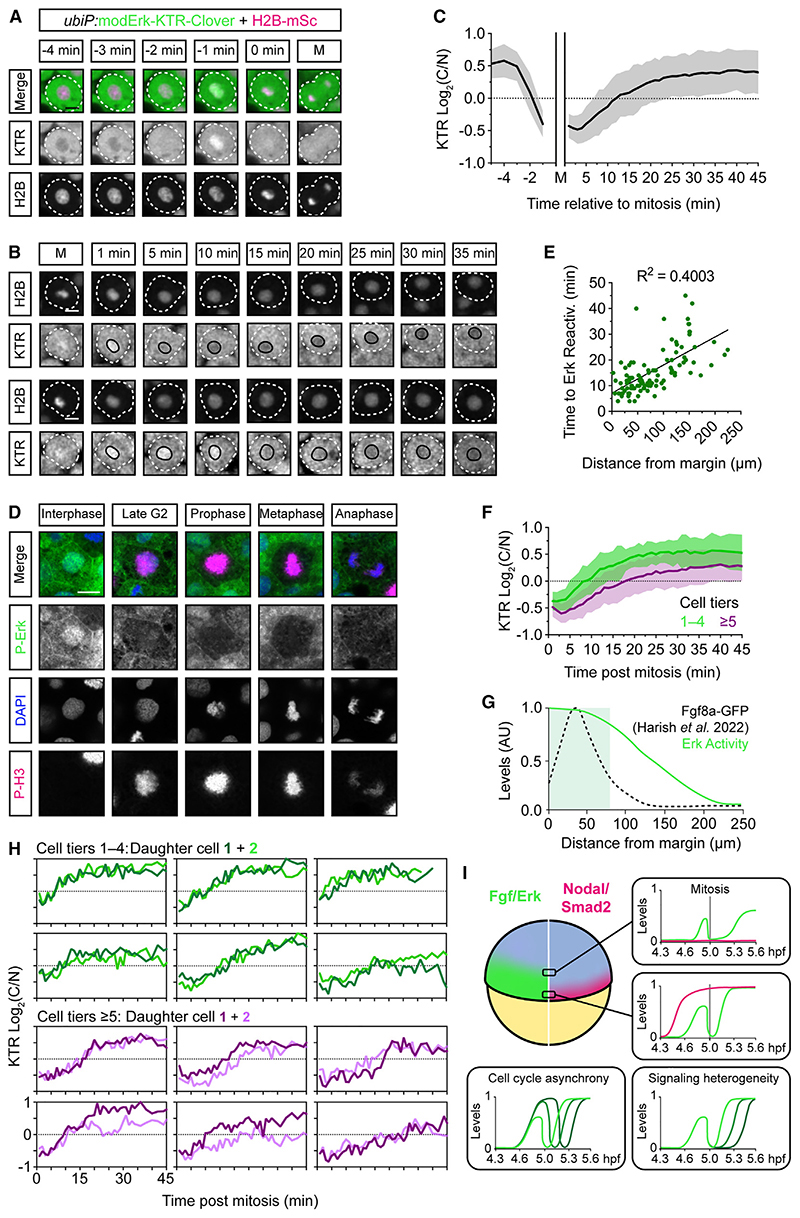
Figure 7 Mitotic erasure induces oscillatory Fgf/Erk signaling dynamics in the presumptive mesendoderm
(A) Representative images of a single mesendodermal cell approaching mitosis.
(B) As in (A) following two cells post-mitosis. Black line labels the nucleus.
(C) Quantification of Erk activityfrom (A) and (B) following mother cells (n = 56 cells) from −5 min before mitosis and daughter cells (n = 110) +45 min after mitosis showing mean ± SD. Nuclear envelope breakdown means the KTR cannot read out Erk activity during mitosis itself.
(D) Representative immunofluorescence images of P-Erk and P-H3 in zebrafish embryos (4.6 hpf).
(E) Quantification of the time to Erk reactivation (log2(C/N) ≥ 0.25) post-mitosis and the distance of each cell from the embryonic margin (n = 110) and fitted with a simple linear regression.
(F) Comparison of Erk reactivation rates from (C) with cells binned based on their initial distance from the margin.
(G) Schematic of the Erk activity gradient, as read out by modErk-KTR, and the extracellular levels of Fgf8a-GFP described in similarly staged embryos (~5.3 hpf).
(H) Single-cell traces of sister cells post-mitosis from (E).
(I) Model depicting how mitotic erasure of P-Erk and its target proteins induces signaling oscillations. Both the rate of reactivation post-mitosis and the final amplitude of Erk activity are sensitive to a cell’s relative position within the Fgf signaling gradient. Coupled with cell cycle asynchrony and variability in reactivation rates, mitotic erasure introduces heterogeneity to Fgf/Erk signaling in the presumptive mesendoderm. Different green lines correspond to P-Erk levels in different cells.
Scale bars, 10 μm.
See also
Reprinted from Developmental Cell, 58(23), Wilcockson, S.G., Guglielmi, L., Araguas Rodriguez, P., Amoyel, M., Hill, C.S., An improved Erk biosensor detects oscillatory Erk dynamics driven by mitotic erasure during early development, 2802-2818.e5, Copyright (2023) with permission from Elsevier. Full text @ Dev. Cell