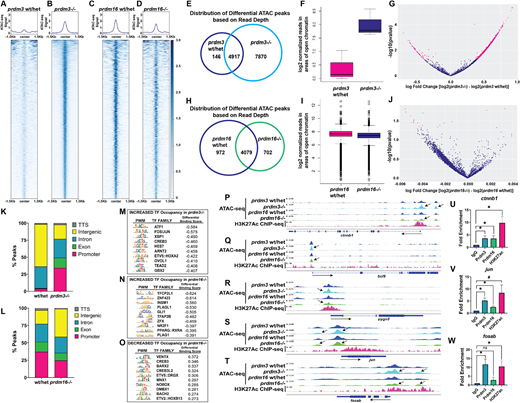
Fig. 5 Prdm3 and Prdm16 alter chromatin accessibility at cis-regulatory regions of Wnt/β-catenin signaling components in cranial NCCs. (A-T) ATAC-seq was performed on sox10:EGFP FACS-sorted cells from wild-type, prdm3−/− and prdm16−/− zebrafish at 48 hpf. (A-D) Coverage heatmaps depicting the differences 1.5 kb upstream and downstream of centered peaks across the genome in controls (A,C), prdm3−/− (B) and prdm16−/− (D) mutants. (E,H) Venn diagrams representing the unique and overlapping differential ATAC-seq peaks based on differences in read depth across two replicates from prdm3−/− (E) and prdm16−/− (H) relative to their corresponding control groups as determined by DiffBind analysis. (F,I) Box plots representing the distribution of normalized read depth concentrations across all significant differential ATAC regions in prdm3−/− (F) and all differential ATAC regions in prdm16−/−. (I) compared with their respective controls. The interquartile range of the box extends from the 25th percentile to the 75th percentile with the line through the box representing the median. Whiskers extend to 1.5× the interquartile range with dots representing outliers. Derivation of these box plots was based a two-sided Wilcoxon Mann–Whitney test computed during DiffBind analysis. (G,J) Volcano plots showing distribution of differential ATAC peaks by fold change and significance in prdm3−/− (G) and prdm16−/− (J). Magenta dots represent those differential peaks meeting the threshold of P≤0.05. (K,L) Annotation of enriched peaks in prdm3−/− (K) and prdm16−/− (L). (M-O) Transcription factor occupancy prediction by investigation of ATAC-seq signal (T±) was performed on prdm3 and prdm16 mutant ATAC datasets. Shown are the top 10 transcription factors with predicted increased occupancy in prdm3−/− (M) and prdm16−/− (N) and decreased occupancy in prdm16−/− (O); P≤E−20. Each table shows the position weight matrix (PWM) for each transcription factor (TF) motif, the transcription factor family, and the differential binding score. (P-T) Tracks showing the distribution of peaks (chromatin accessibility), compared with previously published H3K27ac ChIP-seq datasets (Bogdanovic et al., 2012) from 48 hpf whole embryos, at specific Wnt/β-catenin signaling component target genes that were differentially expressed in prdm3 and prdm16 mutants: ctnnb1 (P), bcl9 (Q), pygo2 (R), jun (S) and fosab (T). (U-W) CUT&RUN paired with RT-qPCR was performed on 48 hpf whole zebrafish embryos with antibodies directed toward Prdm3, Prdm16 and H3K27ac. Enrichment (normalized to IgG) was assessed with primers flanking putative Prdm3- and Prdm16-binding sites near promoter regions (within 1000 bp of the TSS) of ctnnb1 (U), jun (V) and fosab (W). CUT&RUN-RT-qPCR experiments were performed three times; mean±s.d. is shown. *P≤0.05, #P≤0.1; ns, not significant (unpaired, two-tailed Student's t-test).
Image
Figure Caption
Figure Data
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Development