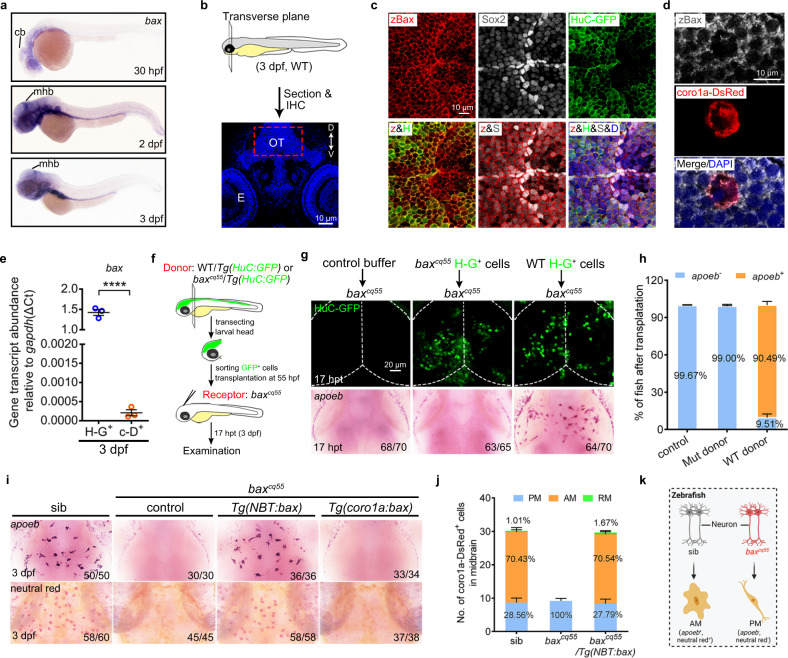
Fig. 2
a WISH of bax in 30 hpf, 2 dpf, and 3 dpf zebrafish embryos. b Schematic illustration of the anatomical position and observational region (red square) in (c) and (d), created with photoshop CS6. c, d Immunofluorescence of zBax (z) with Sox2 (S) and HuC-GFP (H) (c) or coro1a-Dsred (d) in 3 dpf OT region. e qPCR results of the bax transcriptional levels in the sorted H-G+ and c-D+ cells from 3dpf WT midbrains. The data are from three independent experiments. Each dot represents an independent experiment. f The transplantation workflow created with photoshop CS6. g Fluorescent images of H-G+ neurons (top) and WISH of apoeb (bottom) in baxcq55 recipient midbrains at 17 hpt. h The percentage of baxcq55 recipients showing rescued apoeb+ signals in (g). The number in each histogram indicates the average percentage. Data are pooled from three independent experiments. i Representative images of apoeb WISH and NR staining signals in the midbrains of siblings, baxcq55, baxcq55/Tg(NBT:bax) and baxcq55/Tg(coro1a:bax). j The number and percentage of PM, AM and RM in 3 dpf midbrains (n = 6 fish in each group). k Schematic diagram of neuronal Bax regulating microglial maturation, created with BioRender.com. cb cerebellum, mhb mid-hindbrain boundary, IHC immunohistochemistry, D dorsal, V ventral, E eye, OT optic tectum, zBax zebrafish Bax, hpt hours post transplantation, WISH whole-mount in situ hybridization, NR neutral red. The number in each histogram of (h) and (j) indicates the average percentage. Numbers in the right corners in (g) and (i) indicate the counts of embryos with a typical appearance (first number) in the total examined fishes (last number). Error bars, mean ± SEM. ****P < 0.0001, Unpaired two-tailed Student’s t test. Source data are provided as a Source Data file.