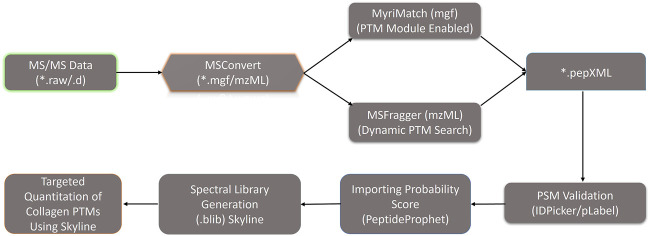
Fig. 2 Optimized dual database search engine-based MS analysis pipeline for the global identification and quantitation of site-specific collagen PTMs from zebrafish heart ECM. Thermo. raw or Bruker. d MS/MS files were initially converted to. mgf and. mzML files (by MSConvert) respectively and searched with MyriMatch and MSFragger to identify the collagen present in the zebrafish heart ECM. For MyriMatch, the subset of identified proteins was used as a second database to perform a PTM module enabled search defining specific sequence motifs for the site-specific identification of collagen PTMs in zebrafish heart ECM. For MSFragger, the PTM searches were conducted directly with the entire zebrafish database. From MyriMatch and MSFragger *.pep.XML output files containing each peptide spectrum match (PSM) were further parsed by PeptideProphet to compute the probability score (0,1). The *.pep.XML output file parsed by PeptideProphet was further imported into Skyline along with all the raw MS/MS files in to generate the spectral library (.blib). This spectral library (.blib) in Skyline was used for the targeted MS1-based extraction of all the PTM modified and unmodified collagen peptide species for each specific site. The area of MS1 area for each peptide for different samples was computed from Skyline.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Front Mol Biosci