Image
Figure Caption
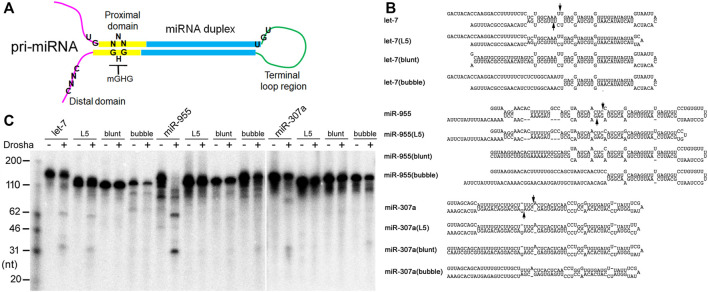
FIGURE 5 Pri-miRNA preferences by Drosha. (A) Schematics of an animal pri-miRNA. The important secondary structural features are shown, along with positions of the UG, UGU, mGHG, and CNNC motifs. (B) Predicted secondary structures of dme-pri-miRNAs and their mutants. Arrows point to Drosha cleavage sites predicted by miRBase. (C) Fruitfly Drosha processing of the dme-pri-miRNAs and their mutants. Positions of DNA markers are shown in the left.
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Front Mol Biosci