Fig 4
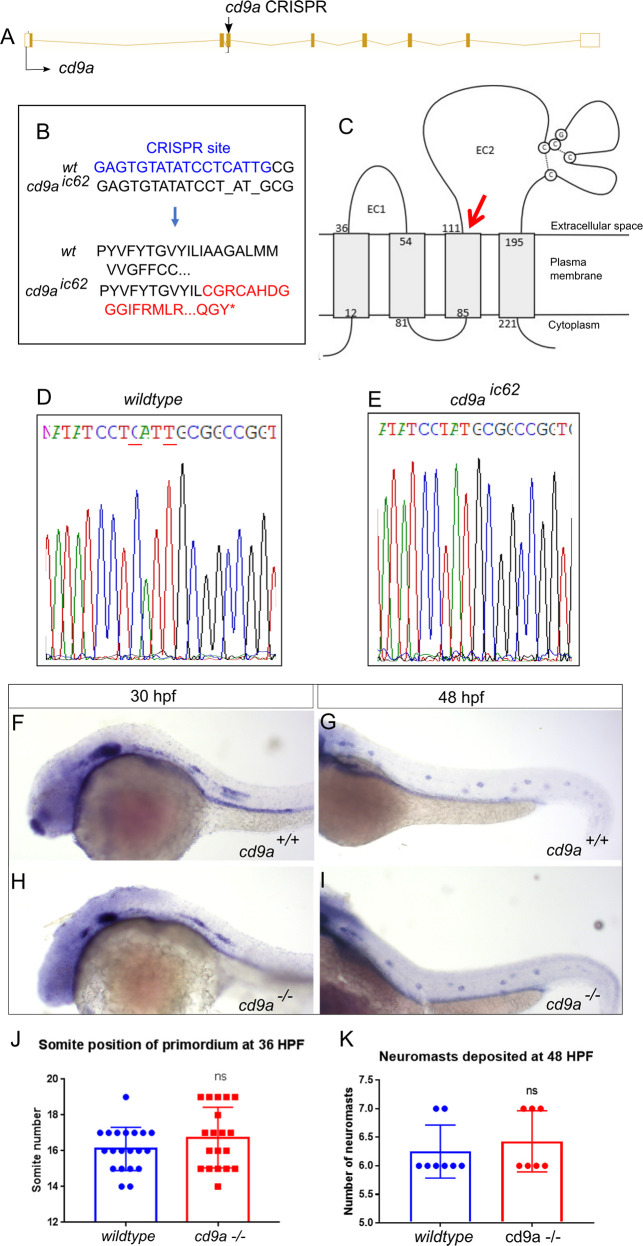
A: Nature of the cd9a mutant allele showing CRISPR site location within the intron-exon structure of the gene. B: The CRISPR target sequence in exon 3 is shown in blue; the 2bp deletion in the cd9a allele is indicated under the WT sequence as dashes. This leads to a frameshift changing codon 62 from ATT (Iso) to TGC (Cys), then 54 aberrant amino acids (red lettering) followed by a stop codon. C: Schematic of the Cd9a protein with location of premature stop codon, indicated by red arrow, at a predicted 115 aa. EC1/2 = Extracellular domain 1/2 D-E: Sequence chromatograms of genomic DNA from (d) WT and (e) cd9aic62 allele. Deleted base pairs are underlined in red. F-G: claudin b expression in WT at (f) 30 hpf and (g) 48 hpf. H-I: claudin b expression in cd9a mutants at (h) 30 hpf and (i) 48 hpf. J-K: Graphs quantifying pLLP measurements in WT and cd9a KOs; (j) migration of the cd9a KO primordium at 36 hpf is similar to WT. (k) There is no significance in number of neuromasts deposited. Significance was assessed using an unpaired t test. N = minimum 7. Bars show mean +/- SD.