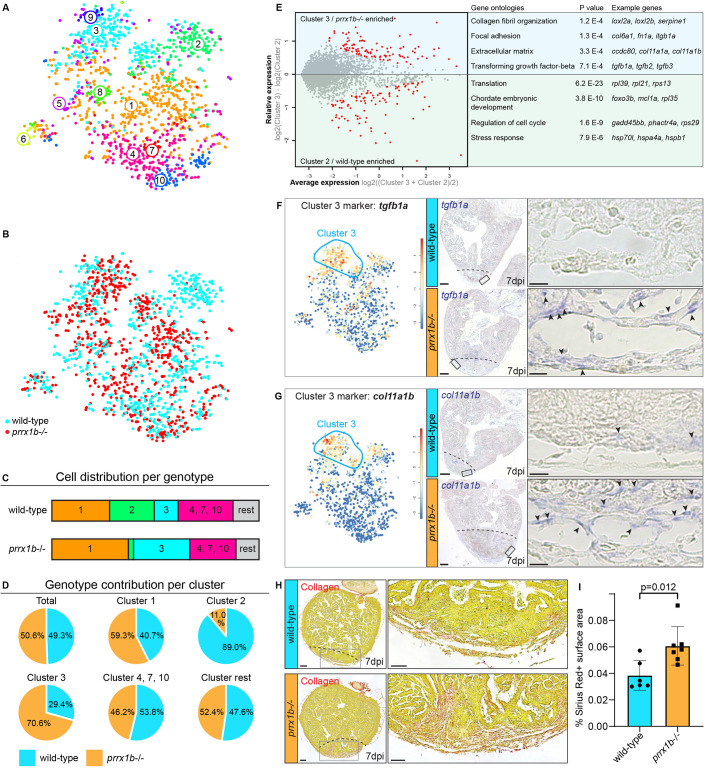
Fig. 4. prrx1b−/− hearts contain excessive amounts of pro-fibrotic fibroblasts. (A) tSNE map of the single-cell sequencing data as shown in Fig. 3C, indicating ten transcriptionally distinct cell populations. (B) tSNE map showing the contribution of wild-type cells (cyan) and prrx1b−/− cells (red). (C) Stacked bar graph showing the relative cell contribution to major clusters in wild-type and prrx1b−/− hearts. (D) Pie charts showing the contribution of wild-type and prrx1b−/− cells per cluster. (E) Differential gene expression analysis using the DESeq algorithm between fibroblast clusters 2 and 3. Enriched genes were selected for either cluster 2 or 3 with a P-value cut-off of <0.05 (red). Gene Ontology analysis was performed using the online tool DAVID. Gene and full Gene Ontology lists are provided in Tables S2 and S3. (F,G) Characterization of cluster 3. Left: tSNE maps visualizing log2-transformed read-counts for genes with high expression in the indicated cluster (circled). Middle: In situ hybridization for the cluster 3-enriched genes in wild-type and prrx1b−/− hearts at 7 dpi. Dashed line indicates injury border. Scale bars: 100 μm. Right: Magnifications of the boxed regions in the injury area with arrowheads pointing to cells with high expression. Scale bars: 25 μm. Three hearts analysed per condition. (H) Sirius Red staining showing collagen in red on sections of wild-type and prrx1b−/− hearts at 7 dpi. Right-hand panels show magnifications of the boxed regions in the sub-epicardial layer and further inside the injury area. Scale bars:100 μm (left); 50 μm (right). (I) Quantification of Sirius Red (collagen) staining in wild-type (n=6) and prrx1b−/− (n=7) hearts showing significantly more fibrosis in prrx1b−/− hearts inside and around the injury area (mean±s.d., P=0.012, unpaired t-test).
Image
Figure Caption
Figure Data
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Development