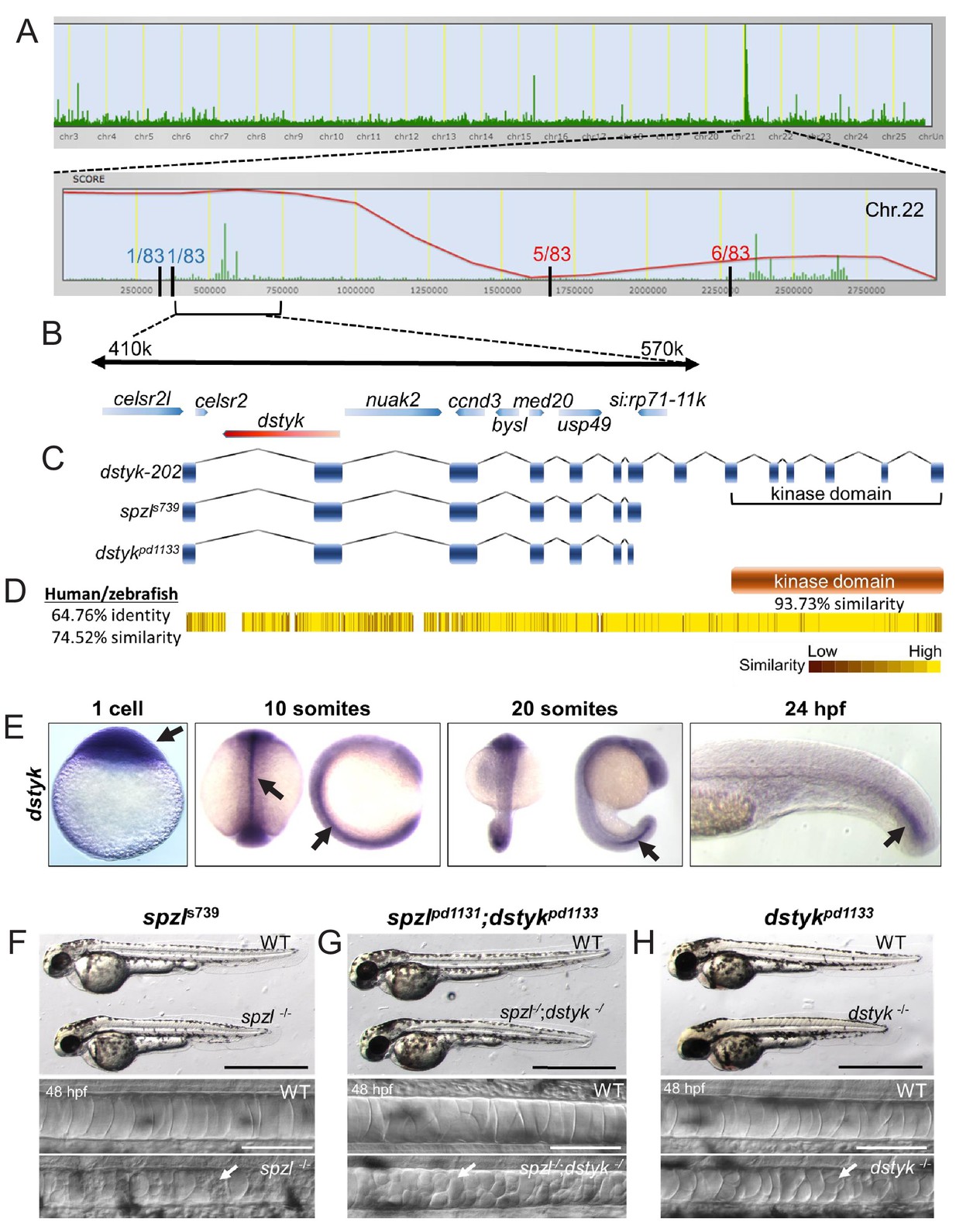
Fig. 3
Loss of Dstyk function causes the spzl phenotype.
( A) Genome view of SNPtrack linkage analysis of exome sequencing data. The spzl locus maps to chromosome 22 (top panel), with a high probability interval (red line) centered around 410–570 Kbps. Recombination events with a panel of 83 mutants at either side of the spzl locus define a ~ 1.2 Mb critical interval. ( B) Genome organization of the critical genomic interval near the spzl locus. A nonsense mutation generates an early stop codon in dstyk. ( C) Cartoon depicting predicted gene product structure of dstyk-202, spzls739 and the Crispr/Cas9 generated dstykpd1133 allele. ( D) Dstyk is a Ser/Thr and Tyr kinase. Heat map depicting amino acid similarity between the human and zebrafish proteins. The kinase domain (highlighted) is 93.73% similar. ( E) In situ hybridization for dstyk expression at the 1 cell stage, 10 somites, 20 somites, and 24 hpf, respectively. Arrows point to highly expressing areas of the notochord. ( F–H) Bright field images of spzl-/-and WT sibling, spzl;dstykpd1133-/- and WT sibling, dstykpd1133 homozygous mutant and WT sibling, scale bars = 1 mm. DIC images of the notochord of 48 hpf embryos. Arrows point to fragmented vacuoles. Scale bars are 100 µm.