Fig. S2
EphrinB1 and EphB3b expression and validation of knock down and knock out strategies. Related to Figure 4.
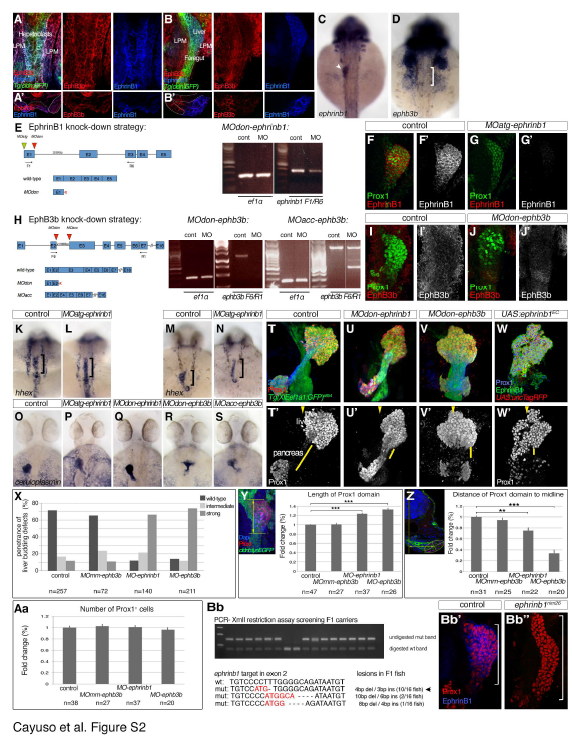
(A-B') Single channel views of immunostainings for EphrinB1 and EphB3b at 22 and 32 hpf shown in Figure 4A-B'''. (C,D) ephrinb1and ephb3b are expressed in the foregut area. At 30 hpf, ephrinb1 is expressed in the liver (arrowhead) and ephb3b in hepatic area (bracket). (E) Schematic representation of ephrinB1 knock-down strategy, with relevant MOs indicated. RT-PCR analysis of MOdon-ephrinB1 embryos at 28 hpf shows a strong knock-down. In addition, a stop codon in the intronic sequence terminates the resulting protein 3 bps after the exon1-intron boundary. Both MO-ephrinB1 produce similar phenotypes (see O-Q), MOatg-ephrinB1 were used for all experiments. (F-G') EphrinB1 expression is abolished or barely detectable in MOatg-ephrinB1 embryos at 32 hpf. (H) Schematic representation of ephB3b knock-down strategy, with relevant MOs indicated. ephB3b maps to linkage group 22 and is partly encoded by EST CT 623350. By 5'RACE, we identified a 2970 bp coding transcript encoded by 16 exons containing all motifs conserved between EphB-receptors, including a N-terminal signal peptide and Ephrin-binding domain. In addition, we found an alternative transcript missing the first two exons, replaced by a N-terminal extension of exon 3 and lacking a signal peptide and a clear transcriptional start site. Its functional significance is unclear and it was not targeted by our knock-down strategy. RT-PCR analysis of MOdon-ephB3b embryos at 28 hpf shows a complete knock-down, indicated by a complete loss of the Ephrin-binding domain-containing exon3. In addition, a stop codon in the intronic sequence would terminate the resulting protein 108 bps after the exon 2- intron boundary. RT-PCR analysis of MOacc-ephB3b embryos at 28 hpf reveals a partial knock-down. This is consistent with the loss of the Ephrin-binding domain-containing exon 3 in a fraction of the transcripts, as indicated by the appearance of a 500bp smaller ectopic band in the MOacc-ephB3b lane compared to controls; control and representative MOacc-ephB3b sample are from different parts of the same gel, as indicated by the separating line. Although, both MO-ephB3b produce similar phenotypes (see O,R,S), the MOdon-ephB3b knock-down is consistently more complete and was used for all experiments. (I-J') EphB3b expression is abolished or barely detectable in MOdon-ephB3b embryos at 32 hpf. (K-N) hhex mRNA expression at 25 hpf reveals that the liver anlage (bracket) forms in MOephrinB1 embryos (L) similar to controls (G), while impaired hepatoblast (bracket) movement into the liver bud in MO-ephrinB1 embryos is apparent at 28 hpf (M,N). (O-S) Two independent MOs targeting ephrinB1 or ephB3b produce equivalent phenotypes, with hepatoblasts positioned more posterior and medial (P,Q and R,S, respectively) compared to controls at 48 hpf (O). Asymmetric hepatoblast positioning is generally more severely disrupted in MO-ephB3b than MO-ephrinB1 embryos. (T-W') Confocal projections reveal liver morphogenesis defects in MO-ephrinB1 (U,U'), MO-ephB3b (V,V'), as well as embryos expressing EphrinB1EC in the context of the entire foregut domain (W,W'). Tg(XlEef1a1:GFP)s854 marks the foregut endoderm, 2F11 the hepatopancreatic ducts and Prox1 the developing liver and pancreas at 54 hpf. Upon loss of EphrinB1 or EphB3b functions hepatoblasts are ectopically positioned in the hepatopancreatic duct domain compared to controls, as indicated by shorter yellow bars, and across the midline (arrowheads; T'-V'). Similar defects are observed upon conditional expression of EphrinB1EC (W,W'). (C,D, K-S) dorsal views, (A-B, F-G', I-J',T-W') confocal projections of ventral views; all anterior to the top. (A’,B’) confocal images of transverse sections, left is oriented to the right. (X-Aa) Quantification and morphometric analyses of liver bud development in MO-ephrinB1 and MOephB3b embryos at 32 hpf. (X) Graph showing the proportion of embryos exhibiting hepatoblast positioning defects in un-injected and MOmm-ephB3b controls, and following EphrinB1- or EphB3b knockdown. In whole-mount preparations the length (Y) and width of the Prox1-domain and its proximity to the embryonic midline (Z) were used to classify the liver budding defects into three categories. (Y) Morphometric measurements showed a significant increase in Prox1-domain length (yellow double arrow) in MO-ephrinB1 and MO-ephB3b embryos compared to controls. (Z) Morphometric measurements revealed a significant reduction in asymmetric displacement of the Prox1-domain from the midline in MO-ephrinB1 and MO-ephB3b embryos compared to controls. Specifically, the distance of the centre of the Prox1-domain to the embryonic midline was determined in transverse sections (yellow double arrow). (Aa) EphrinB1 or EphB3b knockdown does not significantly change the number of Prox1-expressing hepatoblasts; p-values, for all conditions >0.57. (c,e,f) Standard errors are shown; asterisks indicate the p-value for each condition (**, p<0.01; ***, p<0.001). (Bb-Bb'') Stable genetic ephrinb1 mutant was generated using Crispr/Cas9 genome editing. F1 fish were screened for germline transmission of lesions by XmlI digest of amplified target DNA; undigested PCR bands indicate transmission of genetic lesion. Sequencing revealed that F1 fish carry different indels. Intercrosses of carriers with a 4bp deletion and 3bp insertion show a complete loss of α-EphrinB1 staining in homozygous carriers (Bb'') compared to sibling controls (Bb'). A stable line, ephrinb1nim26, was generated from these carriers.
Reprinted from Developmental Cell, 39, Cayuso, J., Dzementsei, A., Fischer, J.C., Karemore, G., Caviglia, S., Bartholdson, J., Wright, G.J., Ober, E.A., EphrinB1/EphB3b Coordinate Bidirectional Epithelial-Mesenchymal Interactions Controlling Liver Morphogenesis and Laterality, 316-328, Copyright (2016) with permission from Elsevier. Full text @ Dev. Cell