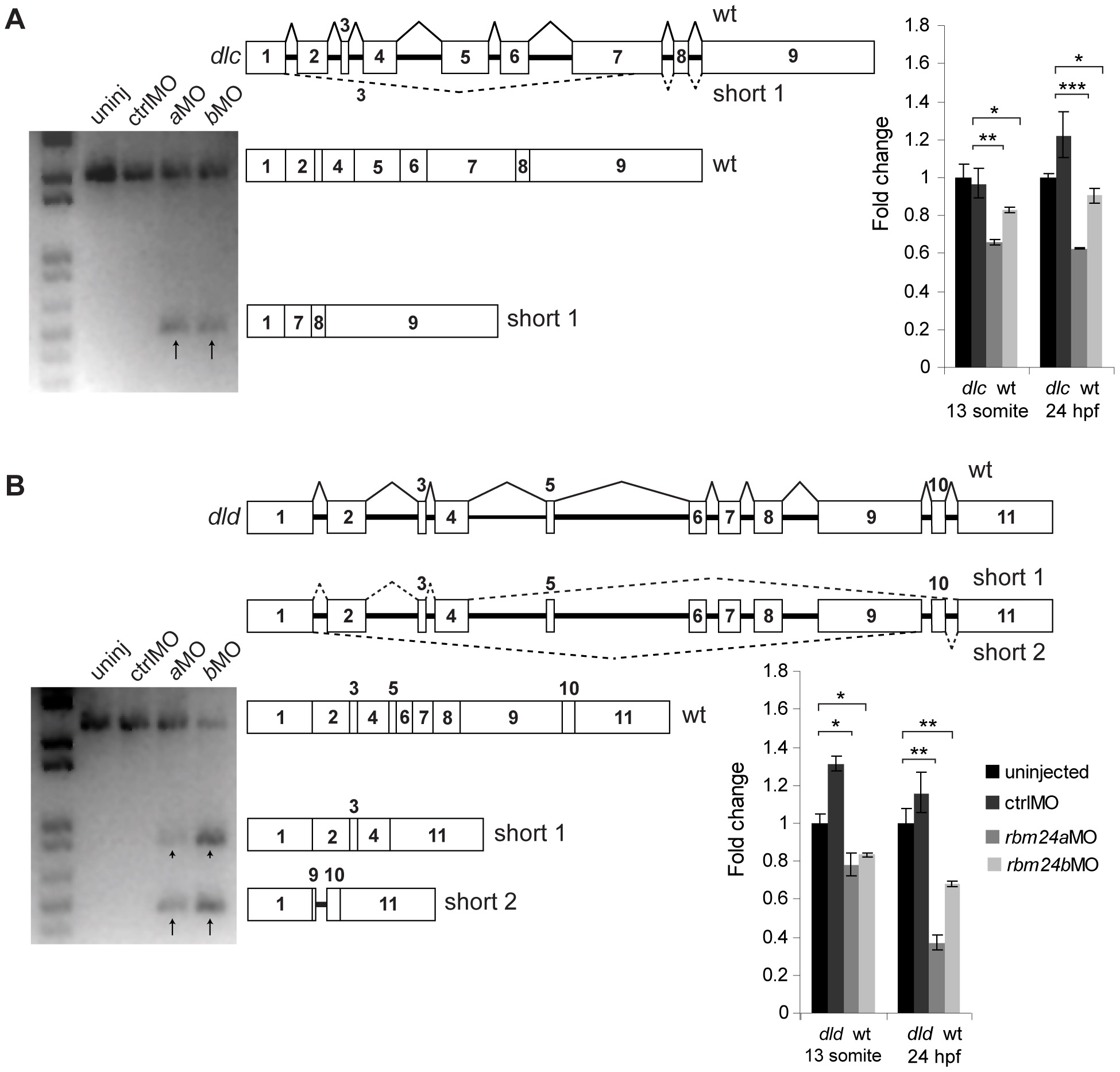
Fig. 4
Aberrant dlc and dld splice forms are detectable in cDNA from rbm24aMO and rbm24bMO embryos.
RT-PCR experiments to amplify dlc and dld mRNA transcripts using total cDNA generated from 13 somite uninjected, ctrlMO, rbm24aMO and rbm24bMO embryos (n = 50 embryos per condition). RT-PCR for dlc transcript yielded a fragment of correct length for all conditions and an additional short fragment for rbm24aMO and rbm24bMO cDNA (A). dlc pre-mRNA, dlc mRNA and dlc short 1 fragment sequences are depicted graphically to scale. RT-PCR for dld transcript yielded a fragment of correct length for all conditions and two additional short fragments for rbm24aMO and rbm24bMO cDNA (B). dld pre-mRNA, dld mRNA and dld short 1 and dld short 2 fragments are depicted graphically to scale. 13 somite and 24 hpf uninjected, ctrlMO, rbm24aMO and rbm24bMO embryos were used to generate cDNA used for qRT-PCR. All transcripts were assayed in triplicate per cDNA sample. Transcript levels were normalized to elfalpha transcript levels and fold change for each transcript is shown as compared to uninjected embryos. An asterisk denotes statistically significant fold changes compared to uninjected. * = p<0.05; ** = p<0.01; *** = p<0.001. black arrows, short dlc and dld fragments; boxes, exons; thick black lines, introns; thin black lines, wild type splicing, dashed lines, aberrant splicing.