Fig. 2
|
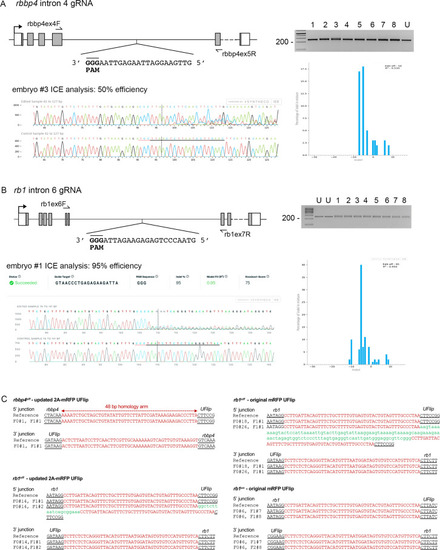
rbbp4 and rb1 intronic gRNA efficiency and F1 UFlip allele junction analysis.
(A) rbbp4 gene model with sequence of the intron 4 reverse strand gRNA. Gel image of PCR amplicons surrounding the target site from 8 Cas9 plus gRNA injected and 1 uninjected (U) embryo. Amplicons from embryo #3 and the uninjected embryo were sequenced and analyzed with Synthego’s ICE software, and indicate 50% indel efficiency at the target site. Plot shows the range and percentage of indels present in the sequences. PAM sequence shown in bold and underlined. (B) rb1 gene model with sequence of the intron 6 reverse strand gRNA. Gel image of PCR amplicons surrounding the target site from eight embryos injected with Cas9 and the gRNA (1-8), and two uninjected embryos (U). Amplicons from embryo #1 and an uninjected embryo were sequenced and analyzed with Synthego’s ICE software, and indicate 95% indel efficiency at the target site. Plots show the range and percentage of indels present in the sequences. PAM sequences shown in bold and underlined. (C) 5’ and 3’ genomic-UFlip integration junctions were PCR amplified from F1 transgenic zebrafish fin clip genomic DNA. The PCR products were sequenced and aligned to the reference sequence expected for a precise integration at the genomic target site. Capitalized red nucleotides represent 48 bp homology arms. Lowercase green nucleotides represent random inserted sequences. |